Cells

Prokaryotes
Domain Archaea
Domain Bacteria
Organelles and Structures
Nucleoid
Concentrated area of DNA (bacterial chromosome)
Store genetic material
Controls cellular activity and reproduction
Transcription and replication of DNA occur here
Plasmid
Area of DNA outside the nucleoid (usually circular)
Carry "nonessential" genes, separately from the chromosome in the cell
Ribosomes
Complexes of ribosomal RNA and protein
Synthesizes proteins
Decode mRNA
Translate to protein

Plasma Membrane
made up primarily of phospholipids
Protects the cell
Provides a fixed environment
Contains cytoplasm
Controls what enters and exits the cell
Selective permeability
Cytoplasm
thick solution enclosed by the membrane
substance in which organelles are suspended
Cell Wall
Rigid shell made of glycoproteins, polysaccharides and pure protein
Provides structure, protection and shape
Peptidoglycan in bacterial cell walls *ONLY*

Glycocalyx
Capsules and slime layers made of polysaccharide and protein
Allow cell to stick or adhere to things
Protect from dehydration
Can protect bacteria from host's immune system

Flagellum
Whip-like appendages
Helps the cell move
Can also act as a sensory organelle
Fimbriae
Short, hair-like projections
Allows cells to stick to things (or eachother)
Pili
Longer projections than fimbriae
Forms channels between bacterial cells
Transfer of DNA from one cell to another
Endospore
A structure that can be created if necessary
Acts as a sort of fail safe for the cell
Preserves genetic material in harsh conditions
Can rehydrate and resume metabolism if/when conditions improve
Eukaryotes
Broken into 4 Kingdoms
Kingdom Plantae
Kingdom Fungi
Kingdom Protista
Kingdom Animalia
can be

Plant Cells
<have these structures<
Chloroplast
Convert Radiant energy into Chemical energy in sugar (glucose) molecules; have thylakoids stacked in grana in the stroma fluid inside the chloroplast membrane.
Cell Wall
Protects cell, Maintains shape, and
Prevents excessive uptake of water.
Made of cellulose, polysaccharides, and proteins.
<Can form channels that connect the cytoplasm of
adjacent cells for transport of molecules<
Plasmodesmata
Both Plant and Animal Cells
<have these Organelles<
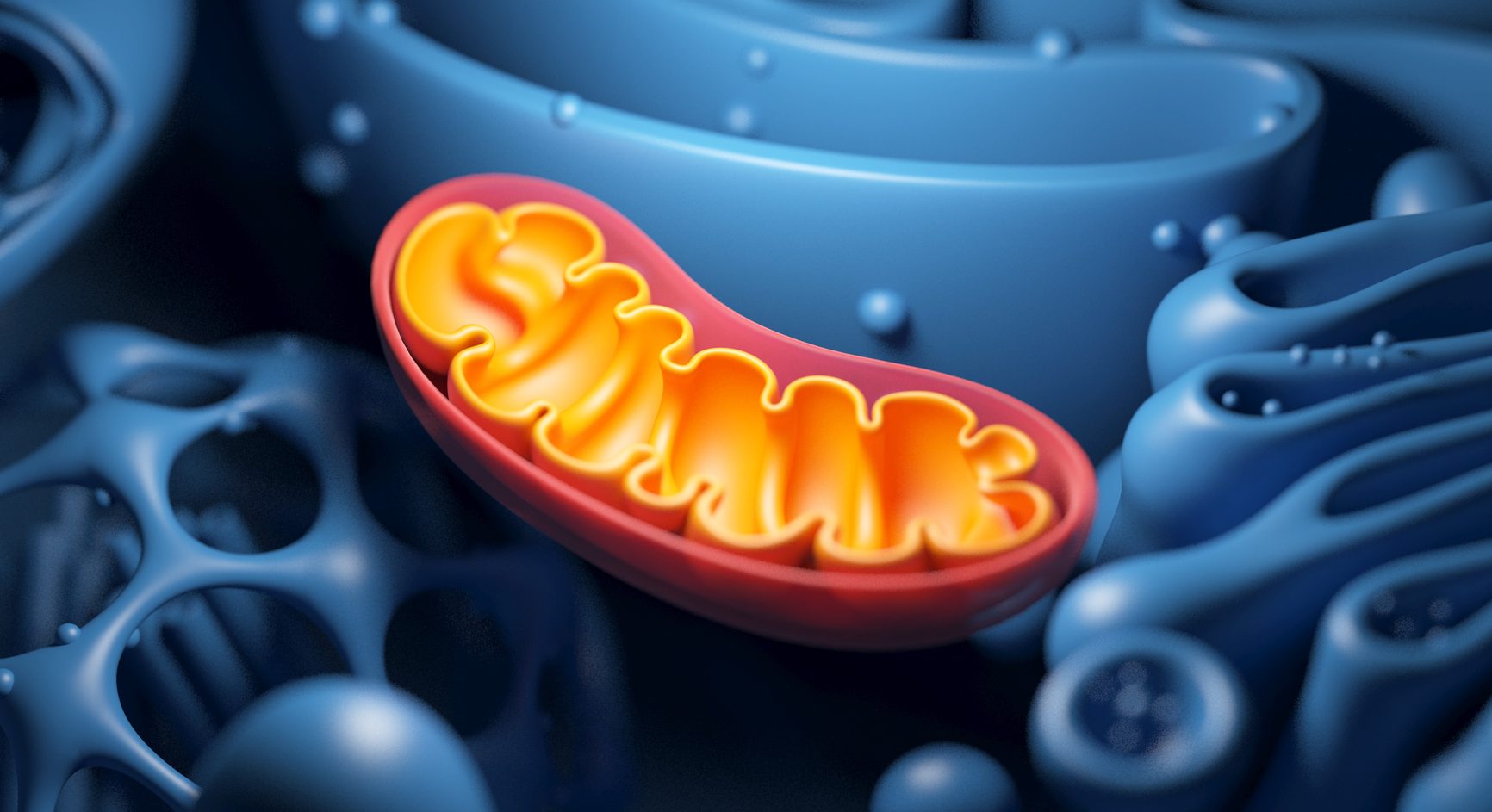
Mitochondria
Produces ATP as energy during cellular respiration
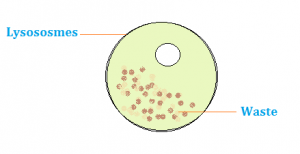
Lysosomes
membranous sac with hydrolytic enzymes that break down macromolecules; made at Rough ER + processed in Golgi body
Function in Phagocytosis (fusing w/ food vacuole and digesting food molecules) and Autophagy (recycling cellular waste)

Vacuole/Vesicle
Selectively transport molecules
^Food Vacuole^
Contractile Vacuole: pump excess water out of cell
Large Central Vacuole: serves as repository for
inorganic molecules

Golgi body
<Consists of<
Flattened membranous sacs (cisternae) with a
cis side (receiving) and trans side (dispatching)
<Function for<
Receiving molecules/proteins from ER; Sorting, Modifying, and Shipping them to be used by other parts of the cell.

Nucleus
Surrounded by a porous nuclear
envelope lined with nuclear lamina.
Contains Nucleolus: site of rRNA synthesis
<"Control Center" containing
genetic information<
DNA made up of Chromatin (chromosomal
units with a histone protein)
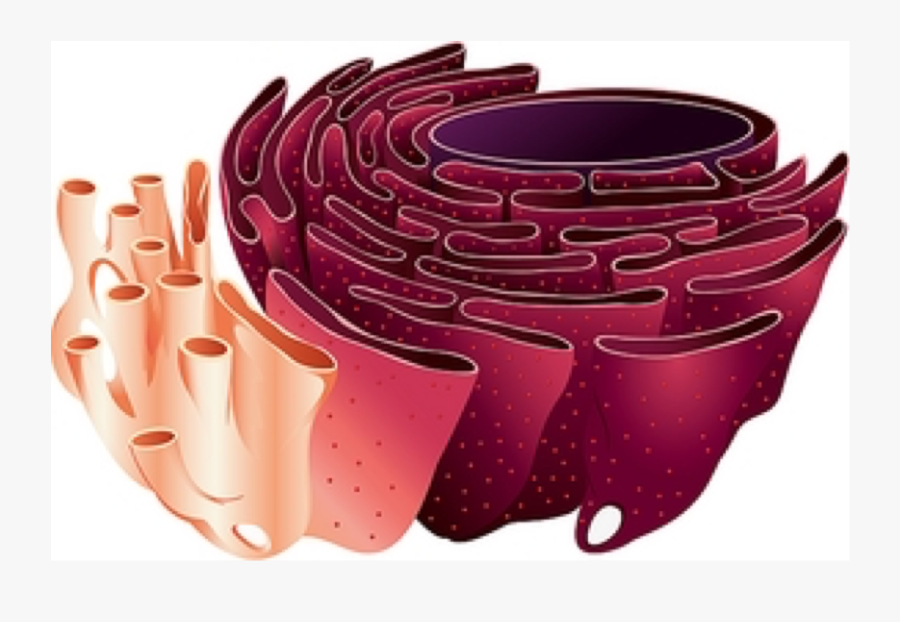
Endoplasmic Reticulum
Smooth ER
Synthesizes Lipids/Carbohydrates ; Detoxifies poisons
Rough ER
Covered with Ribosomes ; Site of Protein Synthesis

Peroxisomes
Contain enzymes that produce Hydrogen Peroxide
and Oxygen, and converts H2O2 into water
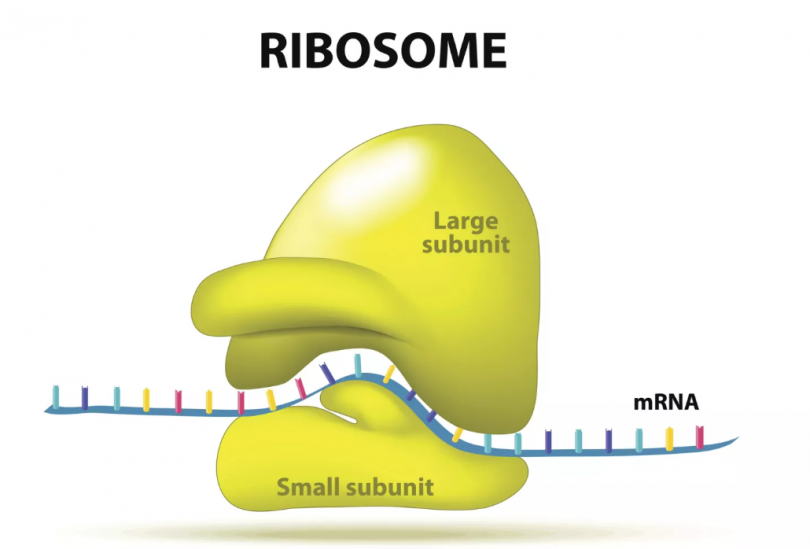
Ribosomes
<Made of<
A small and large subunit (rRNA and protein)
<Serves as<
The site of Protein synthesis

Cytoskeleton
<Supports cell structure/maintain cell shape
+ Involved in cell motility/movement through <
Microtubules
<Come from<
Centrosomes containing a pair of Centrioles made
up of 9 triplet microtubules arranged in a ring (Mostly in Animal Cells; Rare in other Eukaryotic Cells)
<Function for<
Movement of Flagella and Cilia w/
motor protein Dyneins.
Microfilaments
<Made of<
Actin protein molecules
<Function for<
Bearing tension and forming and network
of filaments to maintain cell structure.
Muscle cell contractions with thousands
of actin filaments in a Myosin protein.
Intermediate Filaments
(ONLY in some animal cells)
Each type constructed of
a certain type of molecule
Includes Keratin family
<Function for<
Reinforcement of cell shape +
Fixing position of organelles
Some make up nuclear lamina

Cell Membrane
Encloses organelles/cytoplasm & regulates cell transport
Cell Membrane
Encloses organelles/cytoplasm & regulates cell transport

Animal Cells
<have these structures<
Extracellular Matrix
Made of collagen fibers in a network of
Proteoglycans, Glycoproteins, and other
carbohydrate-containing molecules
Proteins in ECM attach to Integrins
(Cell-surface receptors)
<Regulates cell behavior through
cellular communication by way of<
Tight Junctions
Bind cells' membranes together tightly;
prevent leakage of extracellular fluid.
Gap Junctions
Provide channels for small molecules to pass
between adjacent cells; similar to plasmodesmata
Desmosomes
Anchor cells together in strong
sheets; like in muscle cells
Centrioles
Proteins
Enzymes
Transport
Cell structure
Signaling Molecules
Channels and protein
Defense
Amino Acids
Made of
Amino
Group
Carboxyl
Group
R Groups
Amino Acids

Polypeptide chains

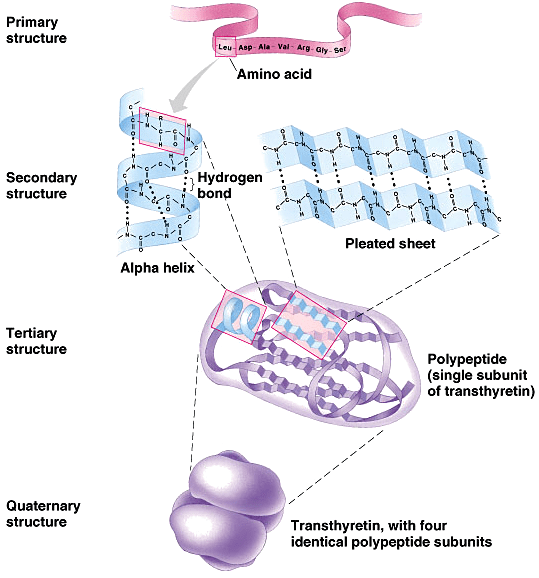
Protein Structures
Primary Structures
Bonds the C-N terminus of Amino acids


Secondary Structures
Side chains of amino acids are attracted to each other through hydrogen bonds
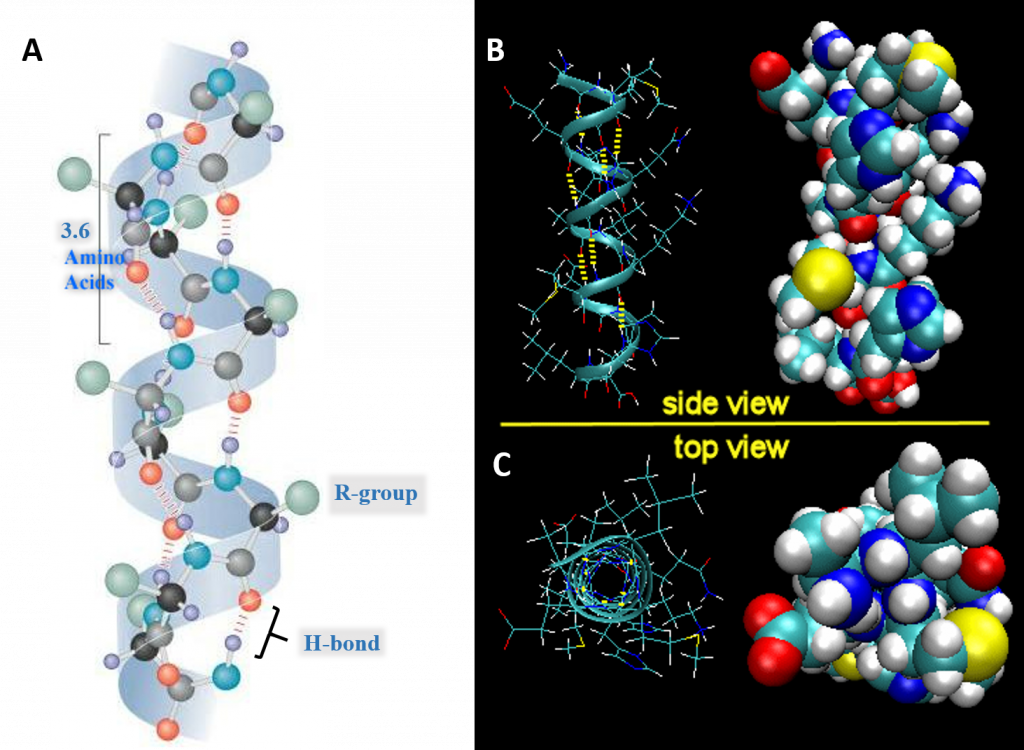
Alpha Helix
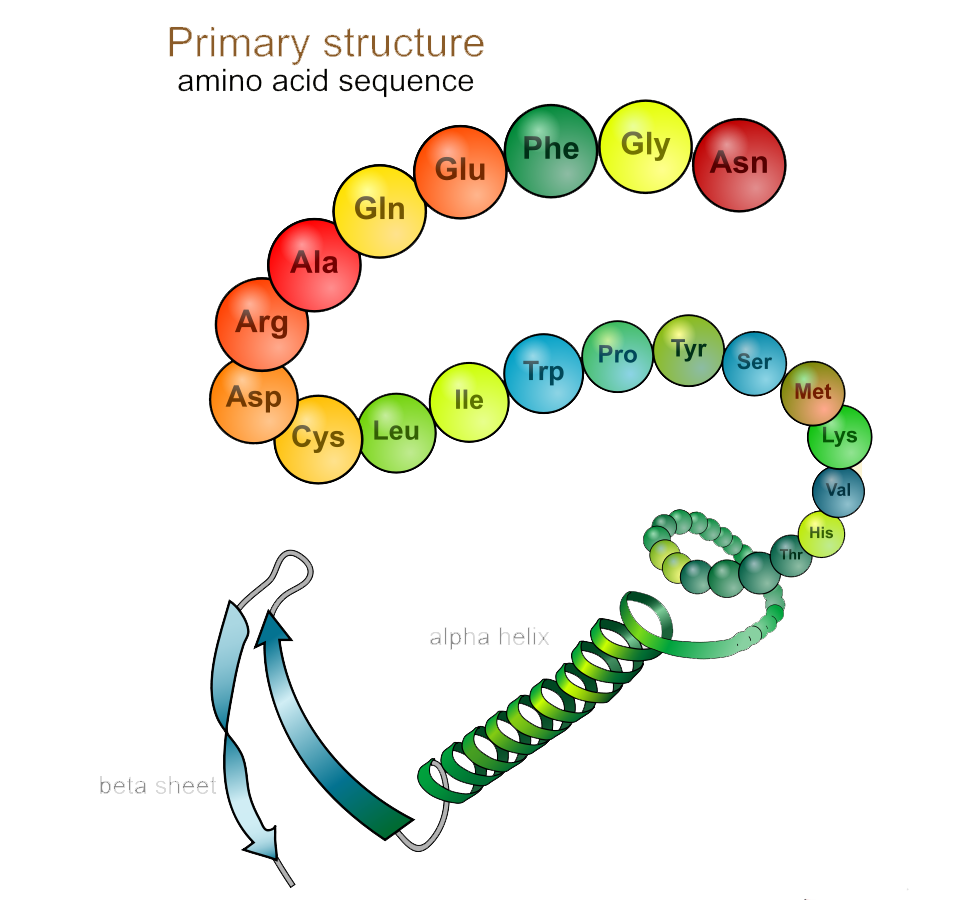
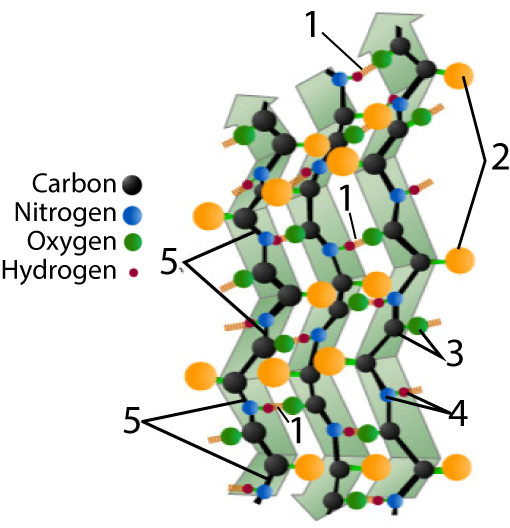
Beta pleated sheets
Tertiary Structures
Through interactions between R groups
Interact through hydrophobic interactions
and vander waals bonds
Interact through hydrophilic interactions
and hydrogen bonds
Disulfide bridges are the only covalent bond between
R groups
Interact through ionic bonds
Quaternary Structures
When multiple polypeptide chains (subunits) come together. They are are held together by several types of bonds and interchain reactions.

Modes of Transportation Across Membranes
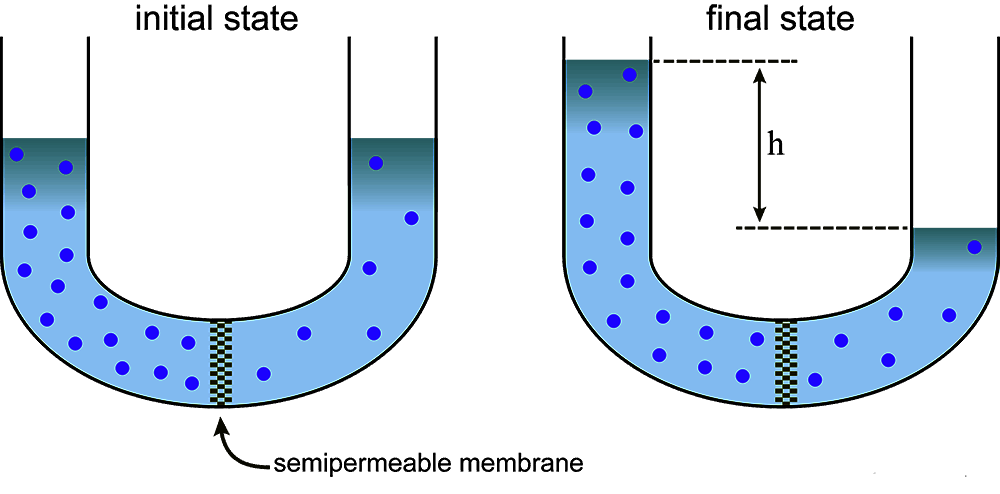
Osmosis
Water moves from concentrations of low solute to concentrations of high solute. Occurs within aquaporins and does not require energy.
Simple Diffusion
Solutes travel from concentrations of high solute to concentrations of low solute. Can transport small non-polar molecules and does not require energy.
Facilitated Diffusion
Facilitated by channel proteins that only allow certain solutes to cross. Moves molecules from high concentrations to low concentrations. Does not require energy and can transport ionic, polar, and large molecules.
Active Transportation
Requires energy and moves molecules against their concentration gradient, from low to high concentration. Capable of transporting ionic, polar, and large molecules.
Phospholipid Bilayer of Membranes
Structure
The phospholipid bilayer is orientated with hydrophilic heads on the outside and hydrophobic tails sandwiched on the inside.
Saturated Tails
Phospholipids with saturated tails have a higher specific phase transition temp. and are therefore more likely to maintain a less permeable viscous membrane.
Unsaturated Tails
Phospholipids with unsaturated tails have a lower specific phase transition temp. and are therefore more likely to maintain a more permeable fluid membrane.
Cholesterol
Cholesterol found between phospholipids within the phospholipid bilayers of animal cell membranes increase the stability and rigidity of the membrane because of its high specific phase transition temperature.
Cell Signaling
Uses ATP
>Comes from>

Cellular Respiration
>Steps Include>

Glycolysis
Occurs in the cytosol. A 6-carbon glucose molecule is converted into TWO 3-carbon pyruvate molecules to be used in Pyruvate Oxidation. ATP is produced through substrate-level phosphorylation. ONLY source of ATP if no O2 (fermentation)
IMPORTANT STEPS (Step 1 and 3):
1. Hexokinase transfers a phosphate group from ATP to glucose to from glucose 6-phosphate [then step 2: phosphoglucoisomerase converts it into fructose 6-phosphate.
2. Phosphofructokinase transfers a phosphate group from ATP to fructose 6-phosphate, converting it to fructose 1,6-biphoshpate --> Key step for glycolysis regulation.
Inputs:
1 Glucose
2 ATP (Energy-
investment)
2 NAD+
Outputs:
2 Pyruvate
4 ATP (Energy-
payoff)
2 NADH
Net:
2 Pyruvate
2 ATP
2 NADH
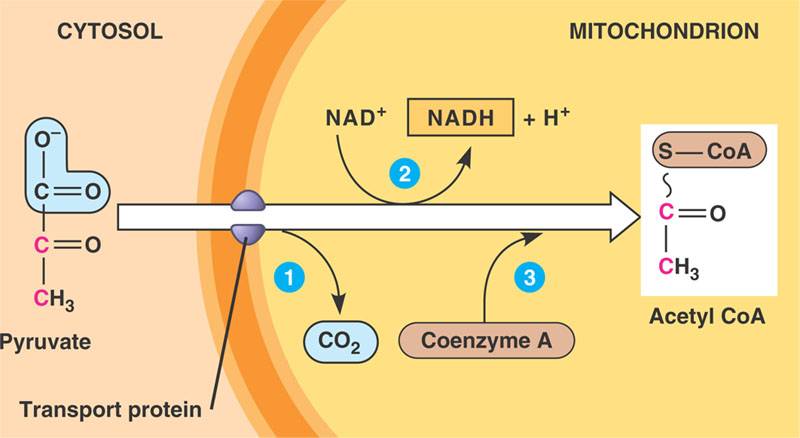
Pyruvate Oxidation
The pyruvate produced from glycolysis enters the mitochondria where undergoes a series of enzymatic reactions. CO2 is removed from pyruvate as it undergoes a redox reaction in which NAD+ is reduced to NADH and pyruvate is oxidized to form Acetyl CoA, which will then go on to be further oxidized in the citric acid cycle
Inputs:
2 pyruvates
2 NAD+
Outputs:
2 Acetyl CoA
2 NADH

Krebs Cycle
Important Steps:
Step 1: Occurs in the the mitochondrial matrix. The Acetyl CoA produced from pyruvate oxidation is converted to oxaloacetate to produce citrate.
Step 3: Isocitrate is oxidized into a-ketoglutarate releasing a CO2 while NAD+ is reduced into NADH. The enzyme catalyzing this step is isocitrate dehydrogenase.
Electrons carriers produced in citric acid cycle is extremely important for the ETC in oxidative phosphorylation
Inputs:
2 Acetyl CoA
3 NAD+
2 FAD+
ADP
Pi
Outputs:
2 ATP
6 NADH
2 FADH2
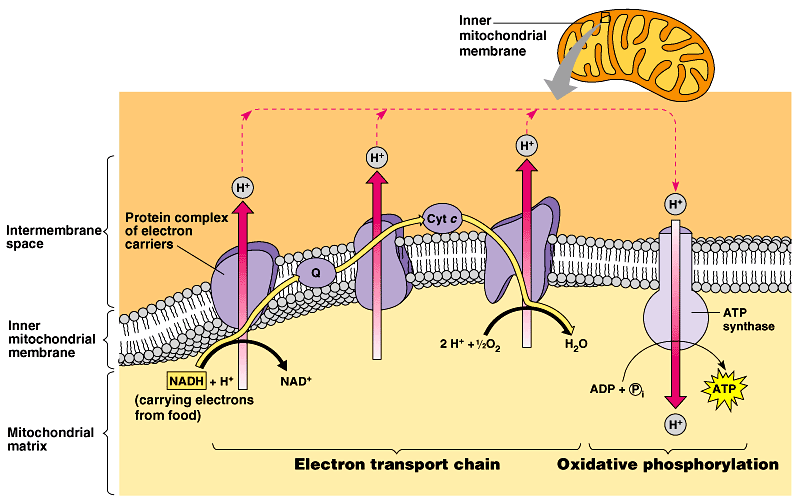
Oxidative Phosphorylation
Occurs in the inner mitochondrial membrane/intermembrane space. Two parts: Electron Transport Chain and Chemiosmosis.
ETC: Electrons from the electron carriers (10 NADH and 2 FADH2) made from glycolysis and the Krebs cycle are transported along proteins in the inner membrane (I, II, Q, III, Cytochrome C, and IV) providing energy to pump protons (H+) out into the intermembrane space against its concentration gradient. The intermembrane space is then highly concentrated with H+ and is low in pH. The electrons are ultimately joined with 2 H+ and 1/2 O2 to produce water (H2O).
Chemiosmosis: The protons in the intermembrane space re-enter the mitochondrial matrix through the enzyme-protein ATP Synthase, providing the energy needed for ADP and inorganic phosphate (Pi) to produce ATP. (endergonic reaction)
Inputs:
10 NADH
2 FADH2
Oxygen
Outputs:
26-28 ATP
Water
Step 1: Signal Reception
<Different types of membrane receptors<
Ion Channel Receptor
The ion channel receptor remains closed, maintaining an ion gradient, until a signal ligand molecule binds to it, activating the receptor.
Once activated the ion channel receptor opens allowing specific ions to flow down their gradient. This is a type of facilitated diffusion.
The rapid diffusion of ions into the cell activates a cellular response.
This can lead to rapidly changing concentrations of different ions in cells, which is especially important to nerve cells
Tyrosine Kinase Receptor
When two separate receptor tyrosine kinase receptors become bound to their respective ligand signal molecules they become dimerized and activated.
The newly formed receptor tyrosine kinase dimer becomes phosphorylated and fully activated by the addition of a phosphate group to each of its tyrosine amino acids. The phosphate groups bound to tyrosine are the products of ATP being hydrolyzed to ADP.
The fully activated and phosphorylated receptor tyrosine kinase dimer can now activate relay proteins that go out into a cell beginning a cellular response.
G-Protein Coupled Receptor
G-Protein Coupled Receptor: The GPCR is a transmembrane protein receptor that is activated by the binding of a specific corresponding signal ligand to the extracellular binding site.
G-Protein: The activated GPCR binds to its corresponding G-Protein, activating it. The activated G-Protein then leaves the GPCR and carries a GTP molecule across the membrane via diffusion to a corresponding enzyme. The activated GTP transporting G-Protein binds to the enzyme where GTP is hydrolyzed to GDP by the loss of a phosphate group, activating the enzyme.
The enzyme in this case is adenylyl cyclase
Step 2: Signal Transduction
Usually occurs across multiple steps involving multiple proteins changing shape, passing the signal to the next protein or molecule.
<Examples include<
Second Messengers: molecules within a cell that relay the signal from the first messenger to the next step in the transduction pathway.
<Examples include<
Cyclic AMP (cAMP)
Cyclic Guanosine Monophosphate(cGMP)
Calcium Ions (Ca2+)
Nitric Oxide
Carbon Monoxide
Phosphorylation Cascade
Step 1: cAMP activates an inactive protein kinase. Then cAMP is converted to AMP by phosphodiesterase
Step 2: The activated protein kinase activates the next protein kinase by taking a phosphate from ATP and adding it to the kinase. Many proteins can be activated during this step, leading to signal amplification.
Step 2a: Protein phosphatases inactivate the protein kinases after they have activated the next protein
Step 3: Eventually, the active protein kinase will phosphorylate a protein that leads to the actual cellular response. This step is the end of the "cascade."
Step 3: Response
Signal transduction leads to the facilitation and regulation of activities in the cell
<Examples include<
Synthesis of enzymes and other proteins
Gene expression
Opening/closing of ion channels
Movement/contraction
DNA
Structure
Sugar-Phosphate Backbone
Deoxyribose Sugar
Phosphate groups
Nitrogenous bases

Purines
Guanine (G)
Adenine (A)

Pyrimidines
Thymine (T)
Cytosine (C)

Nucleotides
Monomer of DNA/RNA
Made up of a deoxyribose sugar, phosphate group and a nitrogenous base
Joined together by phosphodiester bonds to make sugar-phosphate backbone
Nucleosides
Deoxyribose sugar and nitrogenous base (no phosphate group)
Replication
Initiation
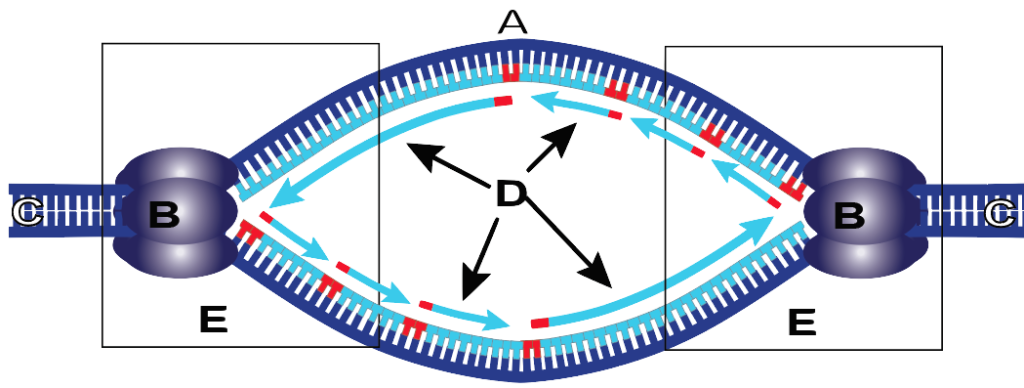
Two strands separate at the ORI sequence and form a replication bubble
<This action is performed by the following proteins<
Helicase: Separates the two strands to form bubble
Single Stranded Proteins (SSB): keep the two strands separate
Topoisomerase: Helps relieve stress caused by unwinding
Primase: Makes RNA primers
Elongation
DNA polymerase III adds nucleotides to the 3' end of the RNA primer and extends it in the 3' direction ONLY
DNA polymerase III with the help of a sliding clamp adds DNA nucleotides to the daughter strand. This polymerization occurs in the 5' to 3' direction.
Eventually, DNA polymerase I will replace the RNA primer with DNA nucleotides so that the new DNA is made only of DNA nucleotides and not RNA nucleotides.
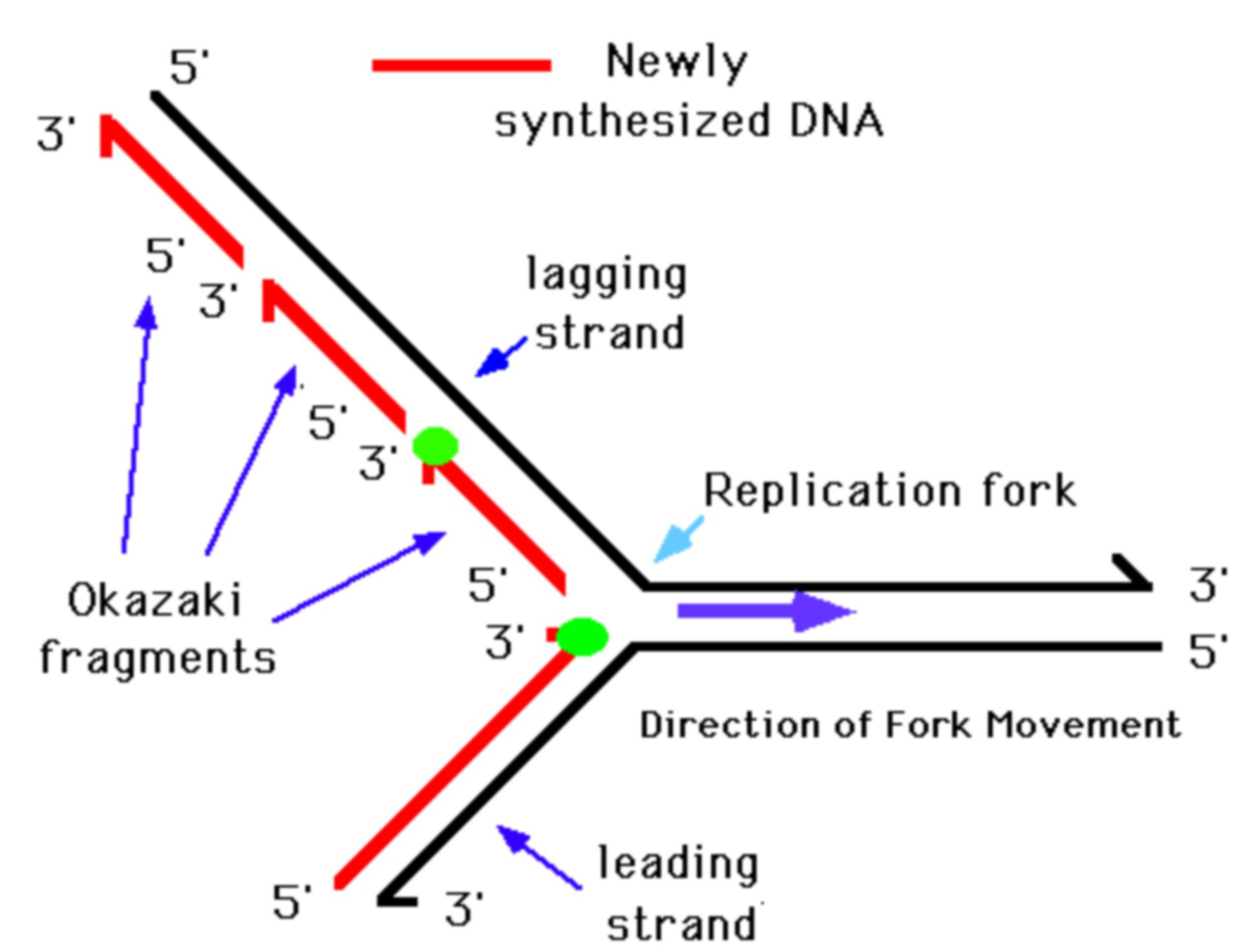
DNA ligase then fills in any gaps between Okazaki fragments in the lagging strand
Termination
Termination occurs when two replication forks meet on the same stretch of DNA
At the end of DNA replication, each of the daughter strands has 1/2 of the parent strand, hence the name semi-conservative replication.
Transcription
A strand of DNA, read in 3' to 5', serves as a template.
PROTEINS/ENZYMES:
RNA Polymerase (II in eukaryotes): synthesizes mRNA strand with a DNA template
Transcription Factors: aid in RNA Polymerase II binding to DNA.
Poly-A Polymerase: adds chain of A's to 3' end of pre-mRNA
Spliceosome (in eukaryotes): removes introns and joins exons in pre-mRNA to form mature mRNA
For Initiation of Transcription:
- Transcription factors bind to Promoter Sequence, which is upstream to the transcription starting point.
- RNA Polymerase II then binds to the promoter sequence and unwinds the DNA strands.
- RNA Polymerase II begins to initiate RNA Synthesis at the transcription starting point (denoted by a +1 on an image) and continues downstream in the 5' to 3' direction.
For Elongation of Transcription:
- RNA Polymerase II continues to synthesize the mRNA in the 5' to 3' direction, adding a new nucleotide to the 3' end of the growing strand.
- RNA Pol II untwists the DNA, exposing 10-20 nucleotides at a time, and the DNA winds back after it has been transcribed.
- The newly made mRNA peels away from the DNA template as it is synthesized.
For Termination of Transcription:
- The RNA Polymerase then reaches a terminator sequence in the DNA that signals for the termination of transcription of messenger RNA.
- Eukaryotes 1) add a G-cap to the 5' end and a Poly-A tail to the 3' end of the product, and 2) form pre-mRNA, which is then processes (introns are removed and exons are joined together) with spliceosomes to form a mature strand of mRNA.
- mRNA is ready to be translated at ribosomes in the cytosol
Eukaryotes
- Occurs in Nucleus
- Transcription occurs before translation
- Forms Pre-mRNA, then Processing forms mature mRNA
- RNA Polymerase recognizes promoter w/ transcription factors and TATA box.
- RNA Polymerase II

Transcription factors + TATA box = Transcription Initiation Complex

40 nucleotides per second in eukaryotes
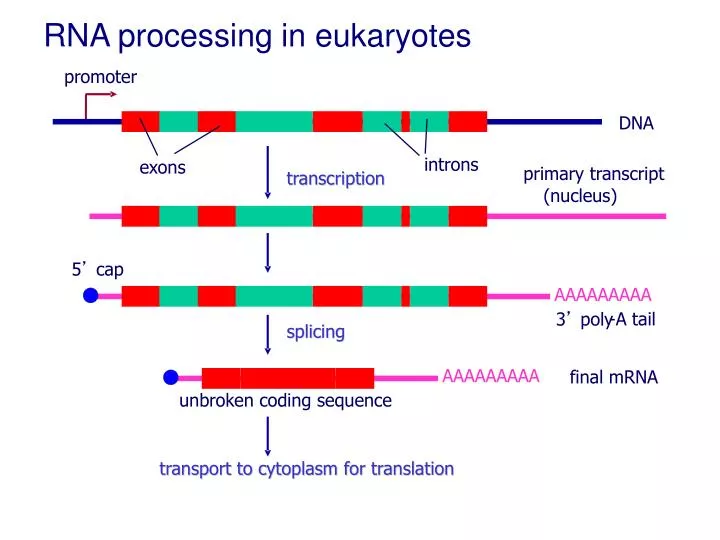
Termination signal is Polyadenylation sequence (AAUAA)
Prokaryotes
- Occurs in Cytosol
- Transcription occurs simultaneously with translation
- Directly forms mRNA
- RNA Polymerase recognizes promoter itself
- RNA Polymerase

Does not use transcription factors or TATA box sequence like Eukaryotes do.
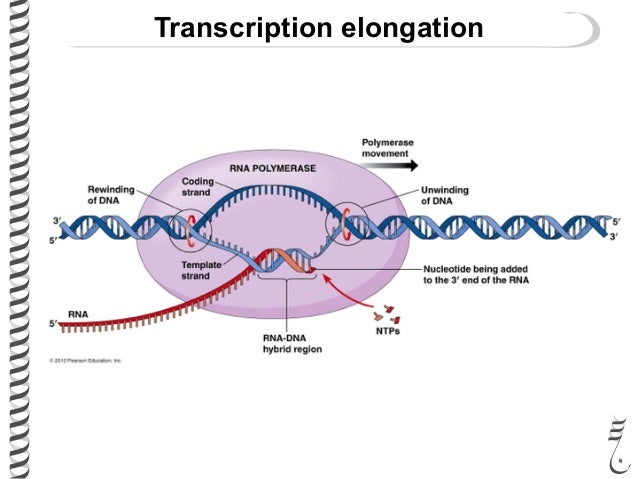
Prokaryotic Transcription
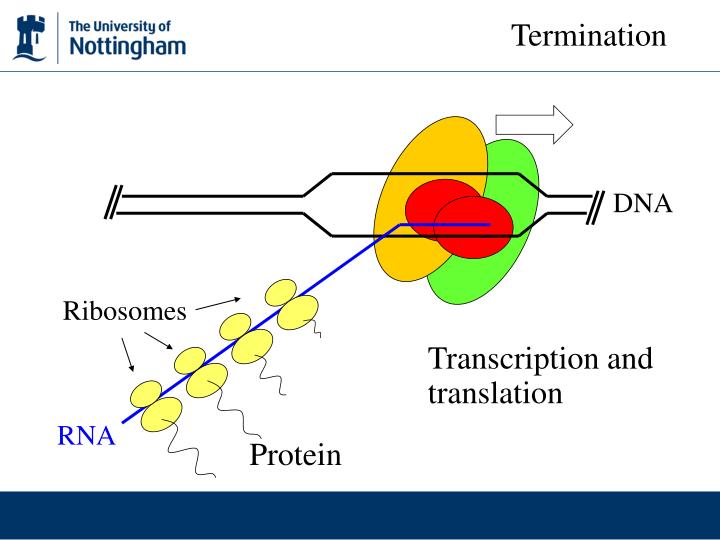
Terminator sequence signals RNA Pol to detach with no further modification.
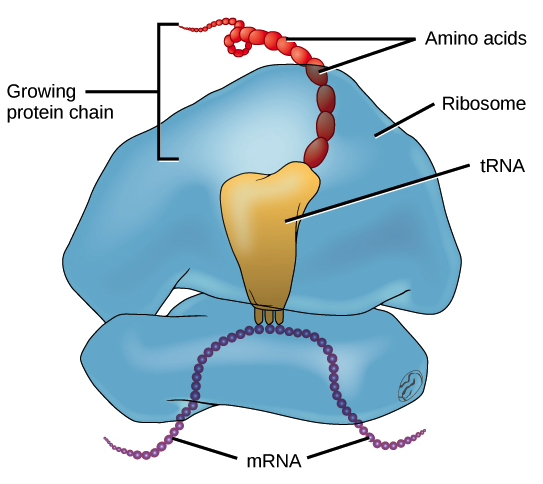
Translation

Eukaryotes
Elongation
- The P site binds the tRNA carrying the first amino acid Methionine (Met)
-Aminoactyl tRNA synthetase is the enzyme that carries
out the matching of the tRNA to specific amino acid
-The A site holds the tRNA carrying the next amino acid to be added to the growing polypeptide chain
-Peptidyl transferase forms peptide bonds between the amino acids from the P site and A site
-the polypeptide is grown in N terminus to carboxyl group direction
- Discharged tRNAs leave the ribosome through the E site.
-Energy required to form the polypeptide chain is provided by
GTP
Termination
Occurs in a response to a stop codon site A , 2 release factors bind to the A sites.
- The nucleotide base triplets UAG, UAA, and UGA (all written ) do not code for amino acids but instead act as signals to stop translation. A release factor, a protein shaped like an aminoacyl tRNA, binds directly to the stop codon in the A site. The release factor causes the addition of a water molecule instead of an amino acid to the polypeptide chain. Thus releasing the polypeptide and disassembling the complex.
Initiation
Components of the Translation Initiation Complex:
-mRNA strands
-tRNA (carries 1st amino acid Methionine)
-small ribosomal subunit
-large ribosomal subunit
The small ribosomal subunit binds to the 5' cap of the mRNA, with the tRNA already attached--> the strand reads downstream until it reaches AUG (start codon)--> the arrival of the large ribosomal subunits complete the initiation complex.

Prokaryotes
Initiation
Components of the Translation Initiation Complex:
-mRNA strands
-tRNA (carries 1st amino acid Formyl methionine)
-small ribosomal subunit
-large ribosomal subunit
The small ribosomal subunit binds the mRNA at specific RNA upstream of the start codon--> tRNA carrying anticodon UAC binds to start codon AUG on mRNA strand--> the arrival of the large ribosomal subunits complete the initiation complex.
Termination
Release factor recognizes stop codon, causes the translational complex to dissociate and the completed polypeptide chain is released.
Elongation
- The P site binds the tRNA carrying the first amino acid formylmethionine (f-Met)
-Aminoactyl tRNA synthetase is the enzyme that carries
out the matching of the tRNA to specific amino acid
-The A site holds the tRNA carrying the next amino acid to be added to the growing polypeptide chain
-Peptidyl transferase forms peptide bonds between the amino acids from the P site and A site
- Discharged tRNAs leave the ribosome through the E site.
-Energy required to form the polypeptide chain is provided by
GTP
Protein Pathways
Upon synthesis at a free ribosome in the cytosol, some proteins will be transported to either the nucleus, a peroxisome, a mitochondria in animal cells, or a plastid in plant cells to fulfill their function without journeying to other organelles.
Some proteins will be temporarily paused during translation by a signal peptide binding SRP. The SRP will then bind at an appropriate receptor and translocation complex on the ER membrane. The signal peptide is then bound to the signal protein complex. The free ribosome has now become a bound ribosome.
Once the ribosome is bound to the ER membrane and the signal peptide is bound to the signal protein complex, the SRP is detached. Translation can now resume with the simultaneous translocation of the polypeptide into the ER.
When the bound ribosome reaches a stop codon the signal peptide is removed from the polypeptide and the polypeptide is released into the ER. Where the polypeptide can fold into a protein.
Proteins formed in the ER will continue their journey to the cis end of the Golgi via transport vesicle. At the trans end of the Golgi the proteins will either be pinched off to form vacuoles or vesicles, that may also become lysosomes, or transported to the cell membrane where they will be secreted from the cell or fulfill a function in the membrane.