Biology 311C
Cell Structures
Eukaryotic Cells
Organelles
Genetic Material:
DNA
stores information
Ribosomes
Bound Ribosomes:
make proteins for membranes
or secretion
Free Ribosomes:
make enzymes & catalyze
sugar breakdown
Vacuoles
Food Vacuole:
cell engulfs food or other particles
Organelles
(membrane bound)
Nucleus
Chromatin:
DNA & protein of chromosomes
Nuclear Envelope:
double membrane
Nuclear Pores:
transport molecules
in & out nucleus
Nucleolus:
site of rRNA
Endoplasmic Reticulum (ER)
Smooth ER:
-lack ribosomes
-synthesize lipids, metabolize
carbohydrates
Rough ER:
-studded w/ ribosomes
-secrete glycoproteins, distributes
transport vesicles
-membrane factory
Golgi Apparatus
Cis Face:
receives cargo from ER
Trans Face:
releases cargo after making changes
Mitochondria:
-endosymbiont
-site of ATP reactions
Double Membrane:
-inner & outer
-inner folded into cristae
-inbetween: intermembrane space
Matrix
-space inside
-steps of cellular respiration
in mitochondrial matrix DNA
-free ribosomes
Peroxisome:
-endosymbiont
-single membrane
-packed w/ enzymes
-detoxify
-hydrogen peroxide to water
Lysosome
(animal cell only)
enzymes inside hydrolyze
biomolecules
Autophagy:
recycles cell's organelles
part of phagocytosis
Chloroplasts
(plant cell only)
-endosymbiont
-site of photosynthesis
Double Membrane
-inner & outer
Thylakoids:
-membranous sacs
-stack = granum
Stroma:
-internal fluid
-has DNA &
ribosomes
Central Vacuole
-plants only
-stores inorganic ions
-lots of water
Cytoskeleton
Microtubules
organelle movement
Centrosome
(animal cell only)
where microtubules
grow out of
Cilia & Flagella:
cell movement
Microfilament
muscle contractions
Amoeboid Movement:
crawling of a cell
cytoplasm movement
Intermediate
Filament
anchor organelles
cell shape
maintenance
nuclear lamina
Cell Wall
-plant cell only
-primary
-secondary
(multiple layers)
Extracellular Matrix
-outside cell
-various proteins
Integrins:
proteins in membrane
that connect ECM to
inside the cell
Cell Junctions
Plant Cell
Plasmodesmata:
-connect neighboring cells
-particles travel freely
Animal Cell
Tight Junctions:
-tightly packed membranes
-no fluid or substance can cross
Desmosomes:
-connection between cells
-some substance can pass through
Gap junctions:
-everything moves
between cells
Prokaryotic Cells
-includes bacteria & archaea domain
Endospores
(not all)
remain viable in
harsh conditions
-original cell copies chromosomes
-surrounds it multiple layers of cell wall
water is removed
metabolism halts
can be rehydrated and viable
Cell Wall:
shapes cell
protection
made of peptidoglycan
in bacteria
Plasma Membrane:
selectively permeable barrier
Organelles
(not membrane bound)
Gas Vacuole:
float in aquatic environments
Nucleoid:
location of DNA
Periplasmic Space:
hydrolytic enzymes
Inclusion Bodies:
storge of carbon, phosphate, etc.
Contractile Vacuole:
pump excess water out
of cell
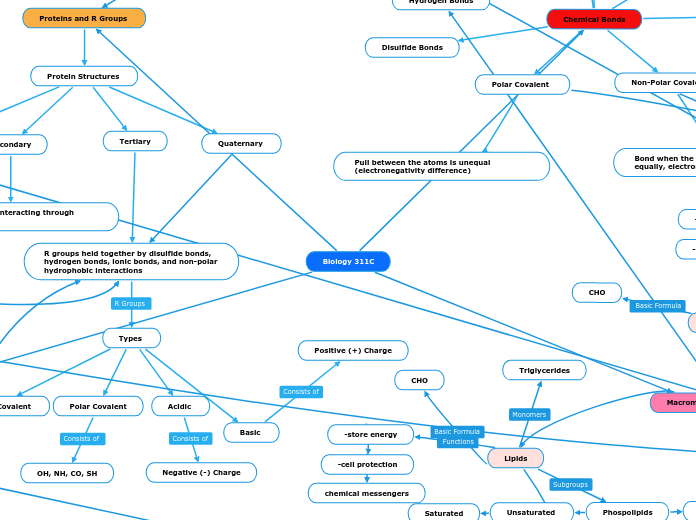
Chemical Bonds
Polar Covalent
Pull between the atoms is unequal (electronegativity difference)
Dipole-Dipole
The force between + end of a molecule (polar) and a - end of a molecule (polar)
Hydrogen Bonds
Is a bond between a hydrogen atom and a very electronegative atom
Van der Waals
When two non polar atoms interact
Ionic Bonds
Bond that includes to oppositely charged ions, + -
Non-Polar Covalent
Bond when the electrons are shared equally, electronegativity is equal
Disulfide Bonds
Proteins and R Groups
Protein Structures
Primary
Amino acids interacting through peptide bonds
Secondary
Main chain groups interacting through Hydrogen bonds
Tertiary
R groups held together by disulfide bonds, hydrogen bonds, ionic bonds, and non-polar hydrophobic interactions
Types
Nonpolar Covalent
CH, ring, H
Polar Covalent
OH, NH, CO, SH
Acidic
Negative (-) Charge
Basic
Positive (+) Charge
Quaternary
Macromolecules
Lipids
-store energy
-cell protection
chemical messengers
CHO
Triglycerides
Phospolipids
Structure
contain four fused rings
Unsaturated
Saturated
Solid at room temperature
hydrocarbon chains of fatty acids are highly packed together and held by hydrophobic interactions
Liquid at room temperature
hydrocarbon chains of fatty acids are not tightly packed together (cis double bonding causes bending)
hydrophilic head, hydrophobic tail amphipathic= having hydrophobic and hydrophilic parts
Carbohydrates
CHO
-provide energy
-spare protein
Monosaccharides
Glucose
C6H12O6
-alpha=carboxyl group at carbon 1 is on top -beta=carboxyl group at carbon 1 is on bottom
Polysaccharides
glycogen (animals), starch (plants),
cellulose (plants), chitin (insects)
Disaccharides
formed when a dehydration reaction joins 2 monosaccharides creating a glycosidic linkage
Nucleic Acids
CHONP
storage of genetic code
nucleotide
DNA
double helix
A and T, C and G
phosphate group, nitrogenous base, pentose sugar (deoxyribose)
RNA
single strand
A and U, C and G
nitrogenous base, pentose sugar (ribose)
Cell Respiration and Fermentation
Glycolysis
Step 1: Hexokinase transfers
phosphate group
from ATP to glucose.
The charge on the
phosphate also traps
the sugar in the cell.
Step 2: G6P becomes Fructose 6 Phosphate
Step 3: Phosphofructokinase
transfers a phosphate
group from ATP to the
opposite end of the
sugar, investing a second
molecule of ATP. Fructose
1,6 bisphosphate is created.
Energy investment:
2 ATP
Energy Output:
4 ATP
2 NADH
2 Pyruvate + 2 H2O
Net Output:
2 ATP
2 NADH
2 Pyruvate + 2 H2O
Happens in the cytoplasm outside of the mitochondria
Pyruvate Oxidation
Pyruvate from Glycolysis is oxidized
Electrons from oxidation are transferred to NAD+ to form NADH
Acetyl Coenzyme A is formed.
Input:
2 Pyruvates, 2 NAD+, 2 CoA
Energy Output:
Acetyl CoA, 2 CO2, 2 NADH
Net Output:
Acetyl CoA, 2 CO2, 2 NADH
Oxygen is not present
Lactic Acid Fermentation
Pyruvate is reduced to form lactate
NAD+ is created and recycled through glycolysis
No CO2 is created
Happens in the cytoplasm
Energy Input: 2 pyruvate, 2 NADH
Energy Output (Net is same):
Lactate, 2 NAD+
Alcohol Fermentation
Happens in the cytoplasm
Energy Input: 2 pyruvate, 2 NADH
Energy Output (Net is same):
Ethanol, 2 NAD+
Pyruvate forms acetaldehyde and releases 2 CO2 in the process
Acetaldehyde is reduced to form ethanol
NADH electrons are reduced creating NAD+
NAD+ goes through glycolysis again
Happens in the matrix of the mitochondria
Oxygen Present
Citric Acid Cycle (Kreb's)
Acetyl CoA interacts with Oxaloacetate to form Citrate.
Citrate becomes isocitrate
Isocitrate oxidizes and NAD+ is reduced to form NADH
CO2 releases when isocitrate is oxidized
Cycle happens twice and NADH and FADH are created
Energy input:
2 Acetyl CoA molecules, 6 NADH, 2FAD
Energy Output:
6NADH, 2 ATP, 2 FADH2, 2 CO2
Net Output:
6NADH, 2 ATP, 2 FADH2, 2 CO2
Happens in the mitochondrial matrix
Fermentation
Oxidative Phosphorylation
Delivery of Electrons by NADH and FADH2
Electron Transport and Proton Pumping
Splitting of Oxygen to form Water
H+ Gradient goes through ATP synthase, creating ATP through oxidative phosphorylation
Energy Input:
10 NADH, 2
FADH2,
*oxygen is the final electron acceptor
Goes through electron transport chain
Energy Output:
26-28 ATP
Net Output:
26-28 ATP
Happens in the proteins in the inner membrane
Plasma Membranes
Phospholipid Bilayer
hydrophilic head
-on outside and inside of cell
hydrophobic tail
-face each other
-middle part of bilayer
Membrane Fluidity
specific phase transition temperature
-above, fluid
-below, rigid
unsaturated hydrocarbon
-tail kinks
-more fluidity
saturated hydrocarbon
-no tail kinks
-more viscous
cholesterol
-reduces movement at
moderate temps
-increases movement at
low temps
-ability to change amount
in membrane to environment
Membrane Proteins
N-terminus
-amino
c-terminus
-carboxyl
Protein Transport
Passive Transport
-no energy used
-with concentration gradient
Osmosis
-diffusion of free water
Aquaporin
Diffusion
-tendency for molecules to
evenly distribute across available
space
Ion Channels
Ungated
-always open
Gated
-open/close in response to stimuli
Stretch-gated
-when membrane
is mechanically deformed
Ligand-gated
-neurotransmitter binds
Voltage-gated
-response to changes in
membrane potential
Facilitated Diffusion
-passive transport aided by proteins
-spread evenly
Channel Protein
-corridor of channel
Carrier Protein
-undergo change in shape
-translocates solute
Active Transport
-uses energy
-against concentration gradient
Sodium-Potassium Pump
Electrogenic Pump
-transport protein that generates
voltage
Proton Pump
Cotransport
-coupled transport
H+/Surcose cotransporter
-active transport driven by
concentration gradient
Selective Permeability
Passes
-small, nonpolar molecules
-small, uncharged polar molecules
Doesn't pass
-large, uncharged polar molecules
-ions
Bulk Transport
Exocytosis
-release contents
Endocytosis
-take in contents
Phagocytosis
-take in food particle
Pinocytosis
-take in fluid
Receptor-Mediated Endocytosis
-solutes bind to receptor site
-form vesicle of bound molecules
Action Potential
-membrane charge flips
Resting State
-step 1
-Na+ and K+ channels
are closed
Depolarization
-step 2
-some Na+ channels
open
Rising Phase
-step 3
-most Na+ channels
open
-cell positive w/
respect to outside
Falling Phase
-step 4
-inactivation of Na+
channels
-K+ channels open
-cell negative again
Undershoot
-step 5
-Na+ closed
-some K+ open
-return to resting state
Metabolism
Catabolic Pathways
Pathways that release energy by breaking down complex molecules into simpler compounds.
Cellular Respiration
C6H12O6+6O2 ———-> 6CO2+6H2O+ Energy
Anabolic Pathways
Pathways that consume energy to build larger, complicated molecules from simpler ones.
Photosynthesis
6CO2+6H2O+Light—>C6H12O6+6O2
Potential Energy
Stored Energy
Example:
-Chemical
Kinetic Energy
The energy associated with the motion of molecules or objects
Examples:
-Thermal
-Light Energy
Thermodynamics
System- Matter within defined region of space
Closed system
Open system
Surroundings- Matter in the rest of the universe
First Law of Thermodynamics-
Energy can be transferred and transformed,
But it cannot be created or destroyed.
Second Law-Every energy transfer or transformation increases the entropy of the universe.
Free Energy
Free Energy Change -
The change in free energy (Delta G) during a chemical reaction is the difference between the free energy of the final state and the free energy of the initial state
Energy changes in chemical reactions
Exergonic
Endergonic
Gibbs Free Energy
Delta G>0
Cannot occur spontaneously. Endergonic Reaction
Delta G=0
A system is at equilibrium.
No change
Delta G<0
A reaction can occur spontaneously. Exergonic
Cell Signaling
-physical contact, local signaling, long distance signaling
membrane receptors (needs help from a second messenger)
G-protein linked receptor
1.) Signal molecule binds to GPRC, once bound, there is a slight alteration in the shape of GPCR which allows the G-protein to bind, this causes GDP to be replaced with GTP which now activates the G-protein
2.) This active G protein can activate a nearby enzyme
3.)nOnce the enzyme is activated, G protein removes the phosphate group from GTP and converts it back to GDP (G-protein has a phosphatase function), this inactivates G- protein
4.) The enzyme activated called Adenylyl cyclase converts ATP to cAMP
5.)cAMP (second messenger) activates other proteins until it leads to a cellular response
Tyrosine Kinase Receptor
1.) signal molecules binds to both receptors which then leads the receptors to bind forming a dimer
2.)Tyrosine is alone so ATP turns into ADP releasing a phosphate group
3.)There are 6 ATP so 6 phosphates and ADP from
4.)These phosphates bind to the 6 tyrosines which leads to cellular responses
Ion Channel Receptor
Ligand Gated Ion
1.) when the ligand attached itself to the receptor the channel opens and allows for specific ions to pass
2.) when the ligand is no longer attached to the receptor the channel remains closed and is inactive
Un-gated Ion Channel
the channel is always opened
signaling molecule (or signal or ligand) and receptor
Intracellular receptors (in cytoplasm and nucleus)
Steroid Hormone
1.) the steroid hormone aldosterone passed thought the plasma membrane (can only pass if it is non polar/ hydrophobic)
2.) aldosterone binds to receptor protein which activates it
3.) the hormone receptor complex enters the nucleus and binds to specific genes that starts the transcription of mRNA
4.) the mRNA is translated into a specific protein
Synaptic Signaling
1.) an action potential arrives, depolarizing the presynaptic membrane
2.) the depolarization open voltage- gated channels, triggering an influx of Ca^2+
3.) the elevated Ca^2+ concentration causes synaptic vesicles to fuse with presynaptic membrane, releasing neurotransmitter into the synaptic cleft
4.) the neurotransmitter binds to ligand-gated ion channels in teh postsynaptic membrane
Gene Expression
Transcription
Location
Prokaryotes:
cytoplasm
Eukaryotes:
nucleus
1. Start
First nucleotide
(+1)
DNA as template
(3'->5')
2. Initiation
RNA polymerases
Prokaryotes:
RNAP
Eukaryotes:
RNA polymerase II
Binds to promoter upstream
(before +1)
Doesn't need a primer
Makes a new strand
(5'->3')
3.Elongation
Transcription factors bind to DNA
(eukaryote only)
RNA polymerase binds
Unwinds DNA
RNA synthesis
(elongates transcript)
DNA reforms in double helix
4. Termination
Prokaryote
-RNA transcript released
-polymerase detaches from DNA
Eukaryotes
-sequence signal to cut
pre-mRNA (AAUAAA)
-release from DNA
RNA Processing
(in eukaryotes)
mRNA
5" End
-modified G nucleotide
-"5' Cap"
3' End
-poly A tail (1-200 A nucleotides)
-made by poly A polymerase
Help:
-exit from nucleus
-prevent degradation
-translation initation
RNA Splicing
Removal of introns
Joining of exons
Alternative Splicing
different combos of exons
are generated
introns help
removal of different ones
make different
proteins
Splicing by Spliceosomes
1. protein snRNA and other proteins
join to become a spliceosome
2. find intron and cut it, intron is recycled
3. join together exons
Translation and Gene Regulation
Translation
For Prokaryotes:
-Simply needs mRNA
-mRNA is already in the cytoplasm when it is created, and it stays in the cytoplasm for translation
mRNA contains code that will help with the formation of proteins
mRNA is made of nucleotides while proteins are made up of amino acids.
Codon chart utilized to show which set of three nucleotides (codons) is coded with which set of amino acids
mRNA in the cytoplasm attaches to free ribosomes:
Prokaryote: 70S ribosomes
Eukaryotes: 80S ribosomes
Aminoacyl-tRNA synthetase matches the correct tRNA with the correct amino acid.
tRNA anticodons bind to the corresponding mRNA codons. (Start codon is AUG)
tRNA first binds to the P site of the ribosome. Then another tRNA binds to the A site of the ribosome (in accordance to the codons on the mRNA) and the amino acid attached to the tRNA in the P site slides over to the tRNA on the A site. While this happens, the tRNA in the P site slides to the E (exit) site of the ribosome and the one in the A site slides to the P site. While this happens, a tRNA attaches itself to the A site. This process continues over and over until the stop codon on the mRNA is reached.
After a protein is created with all of the amino acid bonds, it has two different routes to be a part of the endomembrane system.
To organelles
Protein travels to mitochondria, chloroplast, peroxisomes, nucleus, or just stays in the cytoplasm
To Rough ER (bound ribosomes)
SRP (Signal-recognition particle) attaches to signal peptide
Travels to SRP receptor protein. SRP binds to a receptor protein in the ER membrane. SRP leaves and signal prptide is cleaved by an enzyme in the receptor protein complex.
Protein is released into the rough ER. Several carbohydrate groups are added by enzymes in the ER to the protein, creating a glycoprotein.
Protein travels through a vesicle to the cis Golgi then out of the golgi through the trans Golgi.
Protein then goes from the Golgi to either the plasma membrane, back to the ER, or it gets secreted outside of the cell.
For Eukaryotes:
-Needs mature mRNA
-Mature mRNA travels from nucleus to cytoplasm through nuclear pores.
Players in translation:
mRNA
tRNA
Ribosomes
Amino acids
Amino acyl tRNA synthetase
Peptidyl transferase
Initiation factors
Elongation factors
Release factor
Gene Regulation
In Eukaryocytes
Nucleosomes
Histones +proteins
8 histones in a histone core. Nucleosomes wrap around each histone twice and Linker DNA connects the nucleosomes
H1 Histone is the only one not a part of the nucleosome.
H2A H2B H3 and H4 make up the histone core (2 of each)
10-nm fiber
DNA winds around histones to form nucleosome “beads”
Nucleosomes are strung together like beads on a string by linker DNA
30-nm fiber
Interactions between nucleosomes cause the thin fiber to coil or fold into this thicker fiber
300-nm fiber
The 30-nm fiber forms looped domains that attach to proteins
Metaphase chromosome
The looped domains coil further
Regulation can occur at the level of transcription
Transcription Factors
General:
factors bind to the promoter and regions near the promoter to bring about basal or background level of transcription.
Specific:
activators and repressors bind to distal control elements called enhancers and bring about increased level (activators) or low levels (repressors) of transcription.
Activators and repressors (proteins) are results of cell signaling and are created after transduction.
control elements
Proximal (close to promoter) control elements:
Sequences in DNA close to promoter
Bind general transcription factors
Distal (far from gene they control) control elements
Enhancers
Sequences in DNA upstream or downstream of gene
Maybe close to or far from the gene they control
Bind specific transcription factors (activators/repressors)
In Prokaryocytes
DNA Structure
Experiments
Where is the genetic material, DNA or proteins? - injected S. cells (pathogenic) and R.cells (nonpathogenic) into live mice. When the mice was injected with live S.cells, mixture of heat-Killed S cells and
living R cells the mice died. When injected with living R cells or heat -killed S cells the mice lived
A virus injects cells something to spread so what is being injected? -They infected bacteria with 35S (proteins) bacteriophages and another set of bacteria cells with bacteriophages with 32P (DNA). The bacteria and the bacteriophages were mixed in a tube, after some time they shook the tube to release the bacteriophages from bacterial surface. Then they centrifuged the bacterial cells and looked for the presence of radioactivity – in supernatant and in the pellet. The pellet was blue which meant that DNA was injected into the protein. DNA IS THE HEREDITY MATERIAL
Model of Replication- they showed that after switching bacteria from heavy to light nitrogen and allowing two rounds of replication, their DNA consisted of equal amounts of light and hybrid DNA. Demonstrated that DNA replicated semi-conservatively
The amount of Adenine equals the amount of Thymine. The amount of Guanine equals the amount of Cytosine
DNA is double-stranded, with complementary base pairing.
Replication
Eukaryotic- inside the nucleus prokaryotic- in cytoplasm
Initiation
At the origin of replication, an enzyme Helicase separates the two strands of DNA to form the replication bubble.
SSB or single stranded proteins keep the DNA single stranded. Another enzyme Topoisomerase helps relieve any strain caused by unwinding of the DNA.
Primase – makes RNA primers complementary to the DNA parent strand sequence
The sliding clamp binds to the 3' end, once this happens DNA polymerase lll will add nucleotides only to the 3’ end
Elongation
the DNA polymerase synthesizes a leading strand continuously, moving toward the replication fork. Only one primer is required, the strand is made in one direction 5’ to 3’
In forming the lagging strand, multiple RNA primers have to be laid down and then extended by DNA polymerase III to form short Okazaki fragments.
Another enzyme, DNA polymerase I removes the RNA and replaces it with DNA nucleotides.
Another enzyme called ligase seals any gaps by connecting nucleotides by phosphodiester linkages.
Termination
The 2 replication forks meet and are dismantled, the ends are joined by DNA ligase
2 daughter DNA molecules now have 1 parent strand and 1 new strand (semiconservative)
Protein Transport and Mutations
Destinations of Protein
Nucleus
Peroxisomes
Chloroplast
Mitochondria
Destinations in secretory
Back to the ER
Lysosomes
Membrane Protein
Secretion
Mutations
Mutations also known as mistakes
Can they be corrected? Yes
DNA Polymerase notices the mistakes , and corrects it.
Silent Mutations
Change in DNA= No Change in Amino Acid
Missense Mutations
Change in Change in DNA= Amino Acid Changed
Nonsense Mutations
Change in DNA= Stop Codon is made. Stopping the Sequence.
Frameshift mutations
Change in DNA= Reading Frame is changed